Introduction
Proteins are the functional workhorses of cellular life, carrying out essential tasks that maintain cellular health, growth, and survival. From gene expression to energy production, proteins are central to nearly every biological process. However, their ability to perform these roles is deeply dependent on one critical factor: where they are located within the cell. Some proteins must be in the nucleus to regulate transcription, while others localize to mitochondria to produce ATP, or to the plasma membrane to mediate signaling. These precise placements are directed by intrinsic signals within the protein sequences—like molecular “zip codes”—that guide them to the correct subcellular compartments.
Understanding protein localization is not just a matter of cellular logistics—it’s essential to understanding disease. When proteins are mislocalized, they can lose their function or interfere with the function of others, often resulting in cellular dysfunction. This is especially relevant in cancer biology, where disrupted protein localization can contribute to uncontrolled growth, evasion of apoptosis, and treatment resistance. In the context of radiation therapy, which is commonly used to treat cancer, changes in protein localization after exposure may be part of how tumor cells adapt, survive, and eventually recur. Uncovering these patterns could offer insight into why some cancers resist radiation while others do not.
This project uses machine learning (ML) to predict protein localization based on sequence-derived features, biochemical properties, and public datasets like UniProt. UniProt is a rich, curated resource containing detailed information about protein sequences, structures, and known localizations. By leveraging this data, machine learning models can be trained to recognize patterns—both obvious and subtle—that determine where a protein is likely to reside in the cell. These patterns include amino acid composition, sequence motifs, hydrophobicity, charge, and predicted secondary structures. Some models can also integrate data from protein-protein interaction networks and gene expression levels, offering a more context-aware prediction.
Traditional laboratory methods such as fluorescence microscopy or subcellular fractionation, though accurate, are time-consuming, costly, and difficult to scale. Machine learning presents a faster, more scalable alternative, capable of analyzing thousands of proteins at once and making predictions based purely on sequence data. Modern deep learning approaches, in particular, can extract complex, nonlinear features from protein sequences—often capturing biological signals that traditional models may miss.
Accurate protein localization prediction has far-reaching implications for biomedical research. In cancer, mislocalized proteins have been linked to numerous pathological processes. For example, a mutation that causes a tumor suppressor to be trapped outside the nucleus could render it ineffective, allowing cancer progression. Similarly, resistance to radiation therapy may involve stress-response proteins that translocate to new compartments after exposure, enabling the cell to repair damage or avoid cell death. By building better models for protein localization, we can better understand these mechanisms and potentially identify therapeutic targets to disrupt them.
Just as AlphaFold has transformed the field of structural biology, the future of cellular biology will require models that go beyond static structures and consider the functional context of proteins—when, where, and under what conditions they operate. This project aims to contribute to that future by building predictive models that help uncover how protein localization contributes to disease progression and therapy response.
Although predicting localization is a challenging task due to the dynamic nature of the cell and the complexity of molecular interactions, it is a solvable problem—especially with the right computational tools. By combining deep learning, biological insight, and publicly available datasets, this work seeks to make meaningful contributions to our understanding of cell function and cancer biology. Ultimately, this research could help uncover hidden patterns that drive tumor behavior and open up new avenues for precision medicine.
Topic Explanation
This project focuses on predicting the subcellular localization of proteins using machine learning. Protein localization—the precise positioning of proteins within specific cellular compartments—is critical for maintaining proper cellular function, signaling, and structure. Where a protein resides often determines its role, interaction partners, and biological activity.
Why it matters
- Understanding Function: A protein’s location often defines its function. Predicting localization helps reveal how proteins contribute to specific cellular processes and pathways.
- Disease Research: protein mislocalization is linked to many diseases, including cancer. Improved prediction models can help uncover mechanisms of dysfunction and support the development of targeted therapies.
- Drug Discovery: Knowing where a protein acts enables the design of drugs that target specific compartments, increasing precision and reducing off-target effects.
- Efficiency and Scalability: Traditional localization techniques like microscopy and subcellular fractionation are time-consuming and resource-intensive. Machine learning offers a faster, cost-effective alternative by learning from sequence patterns and large proteomic datasets.
Some examples of important proteins
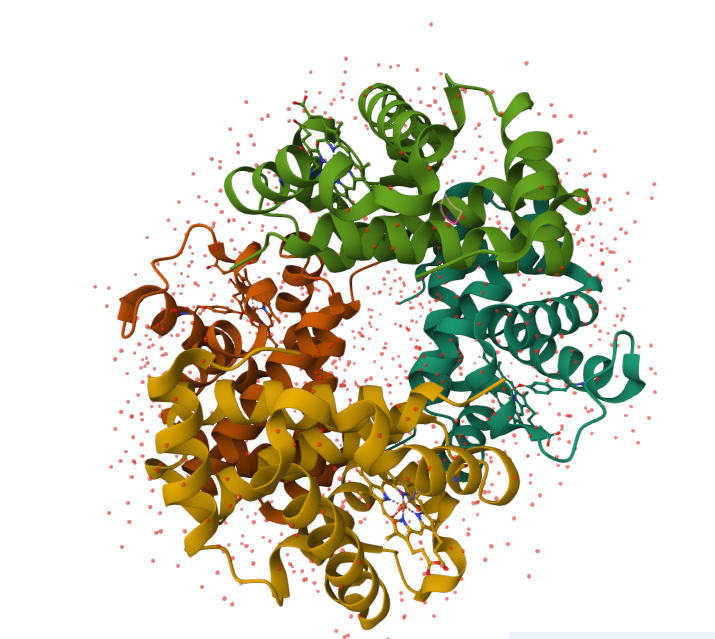
Human p53: protein where its alterd function is most frequently(>50%) associated with cancer.
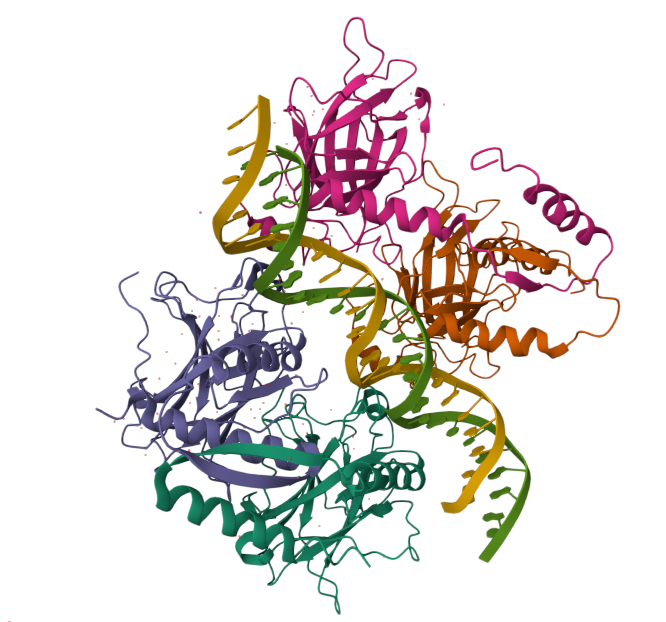
Human hemoglobin: protein contained in red blood cells that is responsible for carrying oxygen to the tissues.
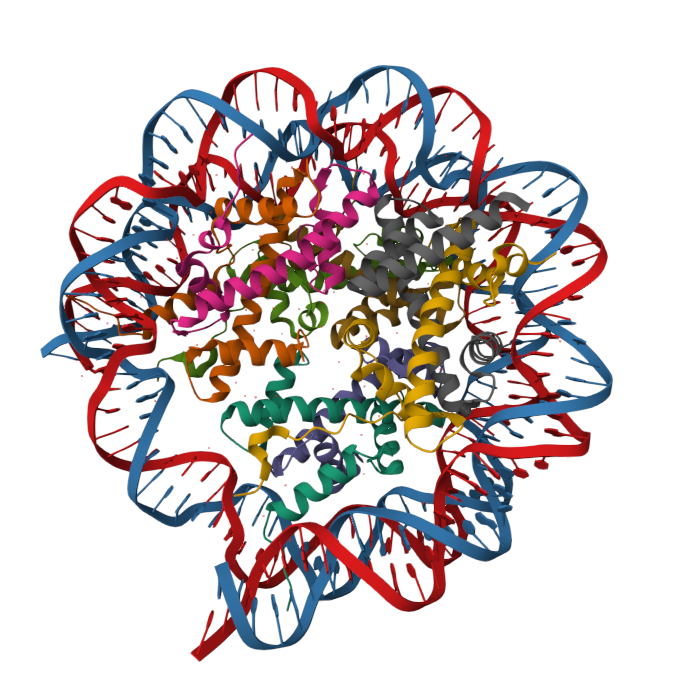
Human histone variant H3.3: Histone proteins are extremely important for packaging DNA in the most efficient way inside cells. They are located in the nucleus of most cells.
10 Questions to be answered
- Is there a pattern in how proteins localize into different cellular compartments?
- How does the amino acid composition vary among proteins in different subcellular locations?
- Is there a correlation between protein length and subcellular localization?
- Are there specific amino acid sequences or motifs associated with certain subcellular locations?
- How does the isoelectric point of proteins relate to their subcellular location?
- Do membrane-associated proteins have distinct characteristics compared to soluble proteins?
- Is there a relationship between protein hydrophobicity and its tendency to form complexes or interact with other proteins?
- What subcellular locations are more easier to predict for?
- What subcellular locations are challenging to predict accurately?
- How is this localization affected by protein modifications?
